YAML calibration documentation¶
This page contains additional information about features and functionality supported by the YAML calibration system
YAML internal structure¶
The native representation of the YAML calibration is as a graph of ‘nodes’ where each node corresponds to either an action or a section containing other nodes. Each node has associated with it a collection of attributes/settings defined for that node (instructions), and a collection of attributes inherited from all parent nodes (context). The atomica.yaml_calibration.build() function takes in the content of the YAML file and loads it into a tree of at.yaml_calibration.BaseNode
instances with a root node called ‘calibration’ at the top level. For example, consider the following YAML file from Tutorial 7:
[1]:
import atomica as at
import pathlib
yaml_file = pathlib.Path.cwd()/'..'/'tutorial'/'T7'/'calibrations'/'T7_YAML_3_repeats.yaml'
print(open(yaml_file).read())
calibration:
repeats: 2
Population calibration:
Match population sizes:
repeats: 5
adjustables: b_rate, mig_rate
measurables: alive
Match deaths:
adjustables: d_rate
measurables: deaths
This file would be loaded into the following tree of nodes:
[2]:
import atomica.yaml_calibration
nodes = at.yaml_calibration.build(yaml_file)
print(nodes)
<SectionNode "calibration" x1>
<SectionNode "calibration" x2>
<SectionNode "Population calibration" x1>
<CalibrationNode "Match population sizes" x5>
<CalibrationNode "Match deaths" x1>
Section nodes have a children attribute that in turn contains others nodes:
[3]:
nodes.children[0].children[0]
[3]:
<SectionNode "Population calibration" x1>
The nodes corresponding to actions each have their own type, with methods that implement the action performed by the node:
[4]:
nodes.children[0].children[0].children[0]
[4]:
<CalibrationNode "Match population sizes" x5>
The context for a node consists of all settings defined in the node’s parents. For example, the ‘Match population sizes’ node inherits the ‘repeats’ context from the parent ‘calibration’ node:
[5]:
nodes.children[0].children[0].children[0].context
[5]:
{'repeats': 2}
The ‘instructions’ for the ‘Match population sizes’ node contains all of the settings that are defined within the node itself - in this case, the adjustables and measurables:
[6]:
nodes.children[0].children[0].children[0].instructions
[6]:
{'repeats': 5,
'adjustables': {('b_rate', None): {'lower_bound': 0.1,
'upper_bound': 10.0,
'starting_y_factor': None},
('mig_rate', None): {'lower_bound': 0.1,
'upper_bound': 10.0,
'starting_y_factor': None}},
'measurables': {('alive', None): {'weight': 1.0,
'metric': 'fractional',
'cal_start': -inf,
'cal_end': inf}}}
The YAML file is executed by sequentially traversing the tree of nodes, and calling the apply() method on each node in turn. The order of execution can be obtained using the walk() method e.g.,
[7]:
list(nodes.walk())
[7]:
[(1, <SectionNode "calibration" x1>),
(1, <SectionNode "calibration" x2>),
(1, <SectionNode "Population calibration" x1>),
(1, <CalibrationNode "Match population sizes" x5>),
(2, <CalibrationNode "Match population sizes" x5>),
(3, <CalibrationNode "Match population sizes" x5>),
(4, <CalibrationNode "Match population sizes" x5>),
(5, <CalibrationNode "Match population sizes" x5>),
(1, <CalibrationNode "Match deaths" x1>),
(2, <SectionNode "calibration" x2>),
(1, <SectionNode "Population calibration" x1>),
(1, <CalibrationNode "Match population sizes" x5>),
(2, <CalibrationNode "Match population sizes" x5>),
(3, <CalibrationNode "Match population sizes" x5>),
(4, <CalibrationNode "Match population sizes" x5>),
(5, <CalibrationNode "Match population sizes" x5>),
(1, <CalibrationNode "Match deaths" x1>)]
This returns a flat list of tuples, where the first item corresponds to the number of times the node has been repeated (which is used when printing progress during execution) and the second item is the node itself.
CalibrationNode functionality¶
Adjustables and measurables settings¶
A calibration node contains adjustables and measurables. Each adjustable and measurable in turn has its own settings. Each adjustable has:
adj_label(required): Adjustable codename (can be found in the framework)pop_name: Population to calibrate (default: all populations)lower_bound: Lowest value the y-factor will be allowed to take (default: 0.1)upper_bound: Highest value the y-factor will be allowed to take (default: 1)starting_y_factor: Y-factor value the autocalibration will start from when running the optimisation algorithm (default: the adjustable’s currenty_factorin the parset)
Each measurable has:
meas_label(required): Measurable codename (can be found in the framework)pop_name: Population to use for calibration (default: all populations)weight: Weight for a particular population (default: weight = 1. This implies that, by default, all populations are weighted equally regardless of size. See the section on setting weights for further details)metric: Metric to be used by the optimisation algorithm (default: fractional)cal_start: Starting year that the calibration will be evaluated for (default:sim_start)cal_end: End year that the calibration will be evaluated for (default:sim_end)
Note that sim_start and sim_end are governed by the project settings and are not set as part of the YAML calibration routine.
When creating a calibration node in the YAML file, it is possible to create the adjustables and measurables using their labels only, using the default values for all other settings. Alternatively, users can specify some or all of the settings. The YAML calibration framework for Atomica supports four ways of setting adjustables and measurables, so as to give users a high level of flexibility. These are:
String format
List format
Dictionary format
Combined format
In general, we can only use one notation within any particular block of adjustables or measurables (with the exception of combined format). However, notation does not necessarily have to be consistent between different calibration blocks, or even between the adjustables and the measurables of the same calibration block.
Below, we will describe each notation and how to use them in a YAML calibration file.
String format¶
The simplest notation is string format, where only parameter names are passed to the adjustables and measurables, like so:
Calibration:
match population sizes:
adjustables: births, mig_rate
measurables: alive
Multiple parameters can be provided, separated by commas. When we use string notation, the optimisation algorithm will perform an autocalibration run using the default settings for adjustables and measurables. If we want to the optimisation algorithm to use specific settings, rather than just the defaults, it is necessary to use one of the other formats.
Dictionary format¶
Dictionary format allows us to explicitly set calibration settings for each adjustable and measurable. We do this by writing the setting names and their values under the relevant parameter name. Each adjustable and measurable is placed on a new line, and their respective settings are also specified on separate indented lines, like so:
Calibration:
Match population sizes:
adjustables:
b_rate:
starting_y_factor: 1.2
mig_rate:
lower_bound: 0.5
upper_bound: 20
measurables:
alive:
cal_start: 2000
cal_end: 2040
We can also specify the same settings for multiple adjustables or measurables at once, by placing them together, separated by commas:
Calibration:
match population sizes:
adjustables:
births, mig_rate:
lower_bound: 0.5
upper_bound: 20
measurables: alive
List format¶
In list format, as the name suggests, we specify the adjustables and measurables settings in a list. It can be useful as a shorthand of dictionary form, since the labels for each setting don’t need to be explicitly written. Instead, we simply write the value of each setting, following the same order as in the Adjustables and Measurables Settings section.
To use list format, place the parameter name and ordered settings values in a list (that is, in square brackets, separated by commas) after the adjustables and/or measurables keyword. The general structure and order to follow are shown below.
adjustables: [adj_label, lower_bound, upper_bound, starting_y_factor]
measurables: [meas_label, weight, metric, start_year, end_year]
Although this might seem like a lot of information for each adjustable and measurable, it is not necessary to include each item every time we use list format – only up to the point where the last setting we want to change is. For example, if we just want to set the b_rate adjustable’s lower bound to, say, 0.2, we only need to list the par_label and lower_bound values in order. Any subsequent settings will retain their default values.
adjustables: [b_rate, 0.2]
measurables: [alive]
However, if we wanted to set values that are at the end of the list order, we need to explicitly specify the default values of all the previous settings. For example, to set the starting_y_factor to 1.2 and the end_year to 2020, assuming the simulation start year sim_start was 2000, we would write:
adjustables: [b_rate, 0.1, 10, 1.2]
measurables: [alive, 1.0, fractional, 2000, 2020]
We can also specify settings for multiple adjustables/measurables at once, by writing a list for each adjustable or measurable, and placing them together in a list of lists. The first example from the previous section on Dictionary format would be written as
adjustables: [[b_rate, 0.1, 10, 1.2], [mig_rate, 0.5, 20]]
measurables: [alive]
In this particular case, it might be most practical to use the dictionary format for the mig_rate, while the b_rate is more concise in list format. List notation may also become convoluted and hard to read if there are parameters to calibrate in the same block. In case like these, we can use the combined format instead, described below.
Combined format¶
Combined format uses dictionary keys, while the values are in list form. This has two main benefits: Firstly, it separates out the parameters in a clear and organised way, which avoids ending up with a dense list of lists containing long series of numbers. Secondly, it allows us to use both the list and dictionary formats under one same adjustables or measurables block.
calibration:
match population sizes:
adjustables:
b_rate:
starting_y_factor: 1.2 ------> dictionary format
mig_rate: [0.5, 20] ------> combined format
measurables: alive
In the above example, the b_rate adjustable settings are in dictionary format, while the mig_rate is now in combined format. When using the combined format, the list of settings is defined in the same order as in the Adjustables and Measurables Settings section. In other words, the order is the same as when using the list format, except we don’t specify the first entry (corresponding to the parameter code name) inside the list, as it is already
specified before the colon.
Calibrating populations¶
The YAML calibration framework allows us to indicate specific populations to calibrate. This can be useful if we wish to calibrate some populations separately, or use different calibration settings for different populations. By default, if only the code name of the adjustable or measurable is provided, a separate copy will be created for every population. To specify the population in any format (except for string format, which does not support populations), the par_name and pop_name must
be placed in a tuple, i.e. in round brackets and separated by a comma, like so: (births, 0-4). Calibrating populations with spaces in the pop_label is supported, and follows the same syntax: (births, 0-4 HIV+). The following are examples of this feature’s usage in all supported formats:
Dictionary format:
adjustables:
(births, 0-4), mig_rate:
lower_bound: 0.5
upper_bound: 20
measurables:
(alive, 0-4):
weight: 0.1
List format:
adjustables: [ [(births, 0-4), 0.5, 20], [mig_rate, 0.5, 20] ]
measurables: [(alive, 0-4), 1.0]
Combined format:
adjustables:
(births, 0-4), mig_rate: [0.5, 20]
measurables:
(alive, 0-4): [1.0]
Meta Y-factors¶
For each parameter, the meta Y-factor is applied to all populations. To calibrate the meta Y-factor and apply the same changes to every population, set the population name to all using the syntax above e.g., (births, all) would set the meta Y-factor for the births parameter
Overriding population specific settings¶
For each adjustable that has been created, the population-specific settings will take precedence over non-population-specific settings. Recall that if no population is specified, this is equivalent to defined adjustables and measurables for each population separately. For example:
adjustables:
births:
lower_bound: 0.5
upper_bound: 20
(births, 0-4)
upper_bound: 10
In this case, births will be adjusted in every population with a lower bound of 0.5 and upper bound of 20, except for the 0-4 population, in which the lower bound will be 0.5 but the upper bound will be 10.
Calibrating transfers and interactions¶
In the case of transfers and interactions, there are two populations involved: the to population, and the from population. The approach is the same as with regular populations, except that we now have two population names in the tuple instead of one:
adjustables:
(aging, 5-14, 15-64):
lower_bound: 0.5
upper_bound: 20
measurables: alive
And in list form:
adjustables: [(aging, 5-14, 15-64), 0.1, 10]
measurables: alive
Calibrating to total population data¶
Usually, our data is structured like below, with each parameter containing several populations, such as age groups.
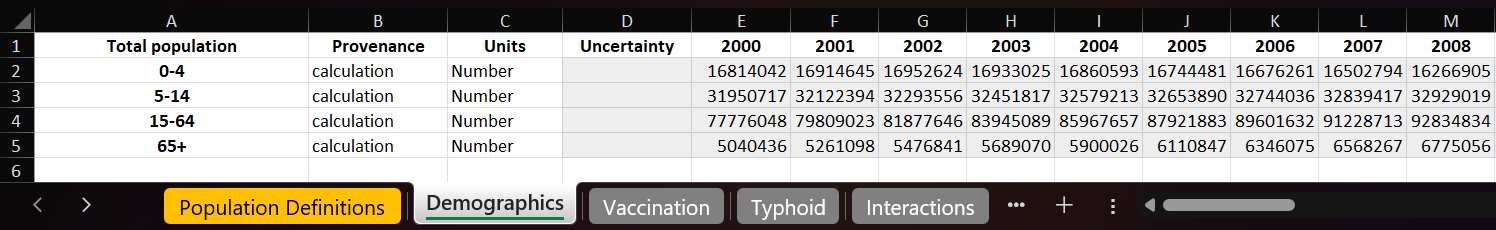
In the YAML file, we could write
calibration
adjustables: b_rate
measurables: alive
Since no population has been specified, all populations will be calibrated.
However, for some parameters, our source data might not be broken down into populations or age groups. In that case, the above YAML file will not work, since there is no data available at the individual population level. What we can do in that situation is to add an extra row to the databook with a population called Total (which is a reserved keyword in Atomica), and explicitly set the population name to Total in the YAML file. If it was our ‘alive’ data that was not broken down by
populations, the databook would look like so
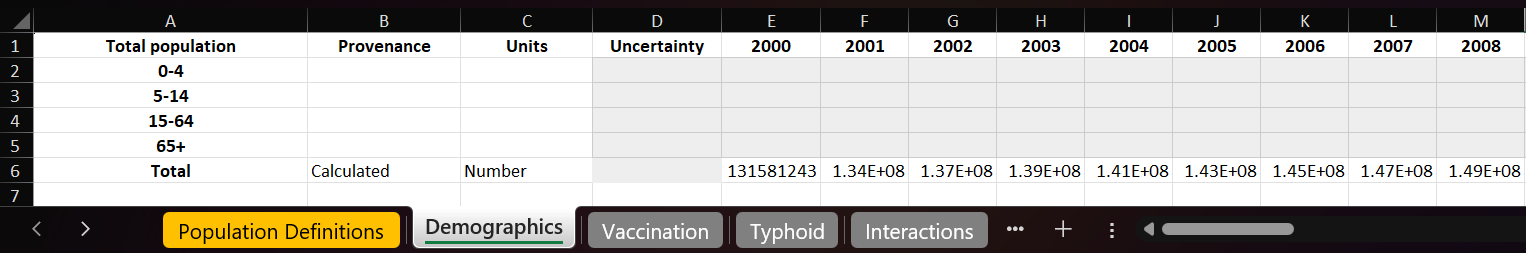
And we would adjust the previous YAML file like so.
calibration
adjustables: b_rate
measurables: (alive, Total)
Measurable Weights¶
In one calibration block, we can include several measurables at the same time, or multiple populations of the same measurable. But say we trusted the data from one measurable’s data source more than another, or wanted to prioritise fitting a particular population – how would we indicate this to the optimisation algorithm?
In the measurable settings, we can set weights for this purpose. The default value for the weight setting is 1.0 which is used for all measurables and populations, which corresponds to giving them each an equal weight, regardless of size. That might be desirable if, for example, we have a key population that is smaller than the other populations – if they were weighted proportionally to size, the small key population might be effectively ignored in the optimisation. However, there might be
cases where we want to do things differently. For example, we could give a key population a higher weight, or we could weight different age bins according to their size. Another reason to use measurable weights could simply be that we trust one data source more than another.
In the following example, we set the 0-4 HIV+ population of the alive measurable to have double the weight than the 0-4 population.
calibration:
match population sizes:
adjustables:
b_rate:
lower_bound: 0.1
upper_bound: 10
measurables:
(alive, 0-4 HIV+):
weight: 2
(alive, 0-4):
weight: 1
Note that the important factor is the proportion between the different weights, not the weight values themselves. That is to say, if we instead set the above weight values to 4 and 2 respectively, the result would be the same.